3437 Mary Sue Coleman Hall, 210 Washtenaw Ave
Ann Arbor, MI 48109
Available to mentor

Janet Smith's research focuses on understanding biological processes through knowledge of the structures of key protein molecules. Early in her independent career, she made major contributions to the understanding of catalysis and regulation in glutamine amidotransferases, phosphoribosyltransferases and photosynthetic proteins by solving and interpreting crystal structures of several proteins of each type. She has also contributed to the development of methods for rapid determination of protein crystal structures, particularly using synchrotron X-ray sources.
A native of Pennsylvania, Smith studied chemistry as a National Merit Scholar at Indiana University of Pennsylvania. Finding biochemistry to be the most stimulating area of chemistry, she continued her study in that field at the University of Wisconsin-Madison where she was convinced of the importance of structure in biology during her research with advisor M. Sundaralingam. Smith then pursued a growing interest in protein structure as a postdoc with Wayne Hendrickson as a National Research Council Research Fellow at the Naval Research Laboratory and as associate research scientist at the Howard Hughes Medical Institute at Columbia University.
Smith established an independent research program in structural biology at Purdue, where she remained as a professor of biological sciences until moving to the University of Michigan and the LSI. She has been a visiting scientist at the European Molecular Biology Laboratory and the European Synchrotron Radiation Facility in Grenoble, France, and a lecturer at numerous international schools on structural biology and synchrotron radiation. She is also a frequent advisor to synchrotron radiation facilities and synchrotron structural biology labs both in the U.S. and abroad.
Smith Lab
-
Center MemberRogel Cancer Center
Our group studies protein structure in order to understand the molecular mechanisms of biological processes and to develop testable hypotheses about function. The lab currently has two research focuses.
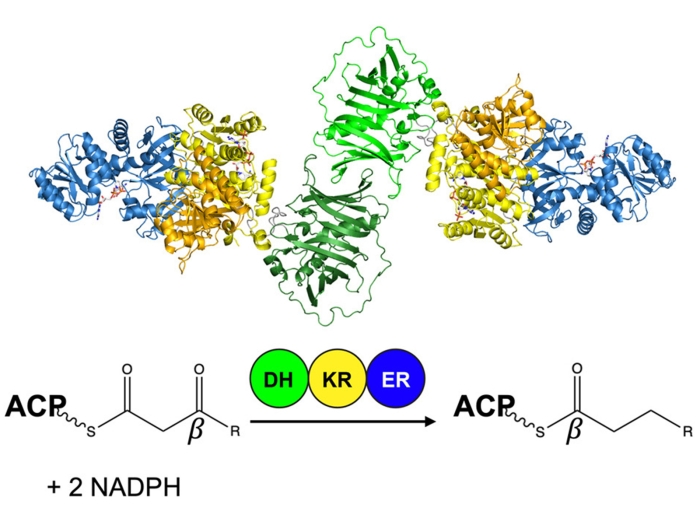
We study the enzymes of natural product biosynthesis with a focus on modular polyketide synthases. One focus of our research is the mechanism of throughput and the substrate/product selecitvity of assembly-line megasynthases. Another is the adaptation of “ordinary” enzymes of primary metabolism to new chemical transformations that enrich nature’s chemical toolbox and have potential as biocatalysts.
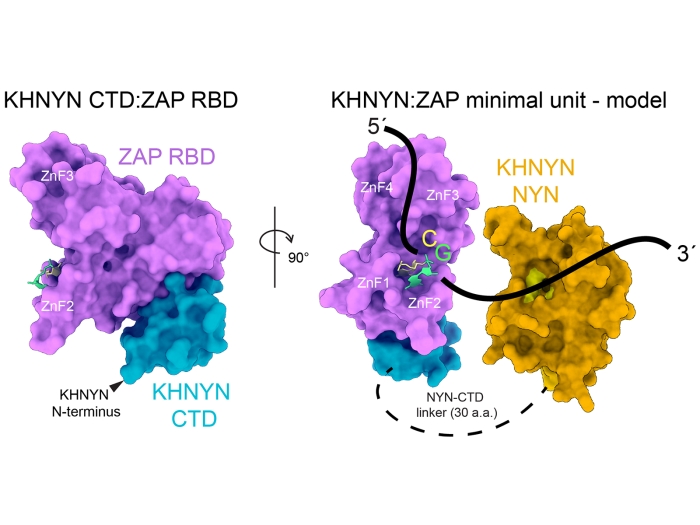
Our studies of the pathogenesis of RNA viruses include both viral and host proteins. Of the host proteins, the zinc-finger antiviral protein recognizes viral RNA in the cytoplasm and targets it for destruction by host nucleases. Other proteins restrict retroviruses by hypermutation of the viral DNA product of reverse transcription. A rapidly evolving viral protein targets the restriction factors for destruction by the proteasome. A multi-functional flavivirus protein helps the virus escape detection by the immune system, increases the infectivity of virus particles, and also has an essential role in viral RNA replication.
-
Rankin MR, Smith JL. Acta Crystallogr F Struct Biol Commun, 2025 Feb 1;Journal ArticleSerendipitous high-resolution structure of Escherichia coli carbonic anhydrase 2.
DOI:10.1107/S2053230X25000068 PMID: 39812168 -
Yeoh ZC, Meagher JL, Kang C-Y, Bieniasz PD, Smith JL, Ohi MD. Proc Natl Acad Sci U S A, 2024 Dec 24; 121 (52): e2415048121Journal ArticleA minimal complex of KHNYN and zinc-finger antiviral protein binds and degrades single-stranded RNA.
DOI:10.1073/pnas.2415048121 PMID: 39693345 -
Bohn JA, Meagher JL, Takata MA, Gonçalves-Carneiro D, Yeoh ZC, Ohi MD, Smith JL, Bieniasz PD. Nat Commun, 2024 Dec 30; 15 (1): 10834Journal ArticleFunctional anatomy of zinc finger antiviral protein complexes.
DOI:10.1038/s41467-024-55192-z PMID: 39738020 -
Rankin MR, Khare D, Gerwick L, Sherman DH, Gerwick WH, Smith JL. 2024 Oct 28;PreprintStructure of a Putative Terminal Amidation Domain in Natural Product Biosynthesis.
DOI:10.1101/2024.10.28.620694 PMID: 39554124 -
McCullough TM, Choudhary V, Akey DL, Skiba MA, Bernard SM, Kittendorf JD, Schmidt JJ, Sherman DH, Smith JL. ACS Catalysis, 2024 Aug 16; 14 (16): 12551 - 12563.Journal ArticleSubstrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
DOI:10.1021/acscatal.4c03637 -
McCullough TM, Choudhary V, Akey DL, Skiba MA, Bernard SM, Kittendorf JD, Schmidt JJ, Sherman DH, Smith JL. bioRxiv, 2024 Jun 20;Journal ArticleSubstrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation.
DOI:10.1101/2024.06.20.599880 PMID: 38948807 -
Mydy LS, Hungerford J, Chigumba DN, Konwerski JR, Jantzi SC, Wang D, Smith JL, Kersten RD. Nat Chem Biol, 2024 Apr; 20 (4): 530 - 540.Journal ArticleAn intramolecular macrocyclase in plant ribosomal peptide biosynthesis.
DOI:10.1038/s41589-024-01552-1 PMID: 38355722 -
McCullough TM, Dhar A, Akey DL, Konwerski JR, Sherman DH, Smith JL. Structure, 2023 Sep 7; 31 (9): 1109 - 1120.e3.Journal ArticleStructure of a modular polyketide synthase reducing region.
DOI:10.1016/j.str.2023.05.019 PMID: 37348494