Joshua Welch, an assistant professor in DCMB, and PhD candidate Jialin Liu received a Chan Zuckerberg Initiative grant for developing two novel computational tools that detect signaling among single cells from spatial transcriptomic data. These tools will provide unprecedented data analysis capability to gain further insights into the communication processes between individual cells.
“A lot of recent work has focused on studying single cells in isolation, but in reality, no cell is an island. We want to detect the molecular messages that cells send each other to understand how cells work together in complex tissue,” said Welch.
Nearby individual cells communicate between themselves through ligands (specialized molecules) that have various degrees of affinity with their receptors. These interactions are important to better understand because they play crucial roles in cell differentiation, tissue homeostasis, immune response, and disease. However, current computational tools that can detect signaling among cell types aggregate cells and do not operate at single-cell resolution. There is also no existing approach that can predict the future states of cell signaling from snapshot data. As a result, our knowledge about individual cell signaling processes is still very limited.
“Deciphering how cells signal to each other during normal development could really help put together the pieces of the difficult puzzle that is tissue formation. This is a key step toward understanding how a few bad cells can cause cascading pathology in disease and how to re-start cell differentiation to allow tissue regeneration,” said Welch.
With support from the Chan Zuckerberg Initiative grant, the U-M team will develop two new computational tools that will help researchers get novel spatio-temporal insights into the processes of intercellular signaling in tissue, at cellular resolution. The first tool, called CytoSignal, performs a statistical test to identify which cells within a tissue have significant activity for a particular signaling interaction. The second tool, called VeloCytoSignal, is a method for predicting the rate of change for a signaling interaction–whether the strength of a signaling interaction is increasing or decreasing at each tissue location–using as little as one time point.
The key challenge of developing CytoSignal and VeloCytoSignal is how to combine gene expression levels and spatial coordinates to predict which cells are sending or receiving molecular signals. “To tackle this issue,” said Liu, “we came up with a straightforward assumption: a cell-cell signaling interaction occurs when all of the requisite ligand and receptor components are expressed in close spatial proximity. This hypothesis aligns with prevailing biological intuition and enables robust inference of signaling dynamics.”
CytoSignal and VeloCytoSignal will be incorporated into one user-friendly open-source R package.
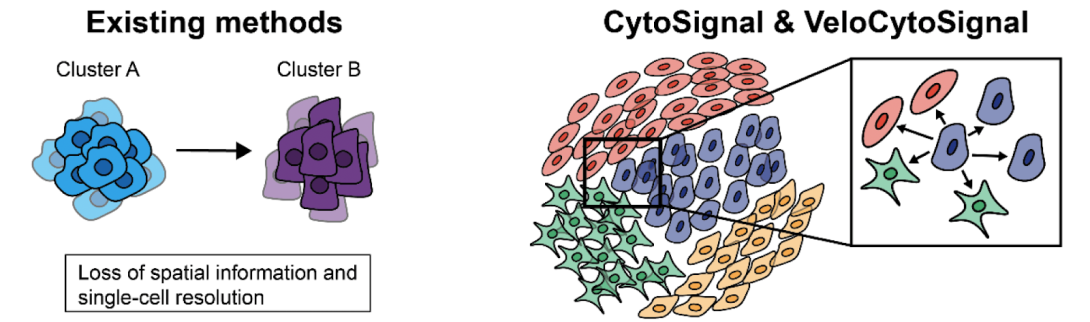
These computational methods are general and therefore can be used with a variety of spatial transcriptomic protocols. This package will take any data modalities as input —as long as they are in the format of a digital expression matrix, such as cell-by-gene matrices, with corresponding spatial coordinates.
Another common challenge in quantitative analysis is that existing tools usually take a long time when analyzing large-scale datasets. The U-M team will develop its package to be an open-source library with great scalability so that it can be easily applied to spatial transcriptomic datasets with large numbers of cells. For example, for a dataset that contains 100,000 cells and 10,000 genes (or features), they expect their tools to finish the whole pipeline in 10 minutes and use no more than 10 GB of RAM using a single CPU core. This high computational efficiency will enable analyses to be performed on personal computers.
The team is grateful for the Chan Zuckerberg Initiative support. This award was granted by the call for projects focused on advancing tools and resources that make it possible to gain greater insights into health and disease from single-cell biology datasets.
The Chan Zuckerberg Initiative was founded in 2015 to help solve some of society’s toughest challenges — from eradicating disease and improving education, to addressing the needs of our local communities. Its mission is to build a more inclusive, just, and healthy future for everyone. This grant from the Chan Zuckerberg Initiative is issued in the second cycle of three cycles, for 18 months.

Associate Professor

PhD Student





