Once again, a University of Michigan (U-M) team of bioinformaticians takes the lead at the prestigious international CASP competition for modeling protein structure from amino acid sequence.
Proteins are the “engines of the cells,” yet the process by which the linear sequence of amino acids folds into a 3-D protein remains incompletely understood. With advances in computational methods and power, it is now feasible to model the 3-D structure of a protein based on its sequence, but major challenges remain for the many scientists who participate in the biennial international Critical Assessment of Structure Prediction (CASP) competition to advance the state-of-the art.
Since 1994, expert participants are invited to submit models for a set of proteins for which the experimental structures are not yet publicly known.
This year, the models submitted by the University of Michigan team founded by Dr. Yang Zhang and led by Dr. Peter Freddolino, a Professor of Biological Chemistry and Computational Medicine and Bioinformatics (DCMB) at the Medical School set a new standard for high-accuracy protein structure prediction. After a dramatic enhancement over the past 2-4 years by Google’s DeepMind AlphaFold, the U-M team took first place in the Multimer and Interdomain Prediction categories, and was again the top-ranked server in the Regular (domains) category according to the CASP assessor's criteria. "The results were assessed as regular protein domain modeling, regular protein inter-domain modeling, and protein complex modeling,” said Dr. Wei Zheng, the major developer of the CASP15 algorithms from the Freddolino lab at DCMB. “Our modeling results were extremely strong.” Qiqige Wuyun (Michigan State University) also participated in the Michigan team.
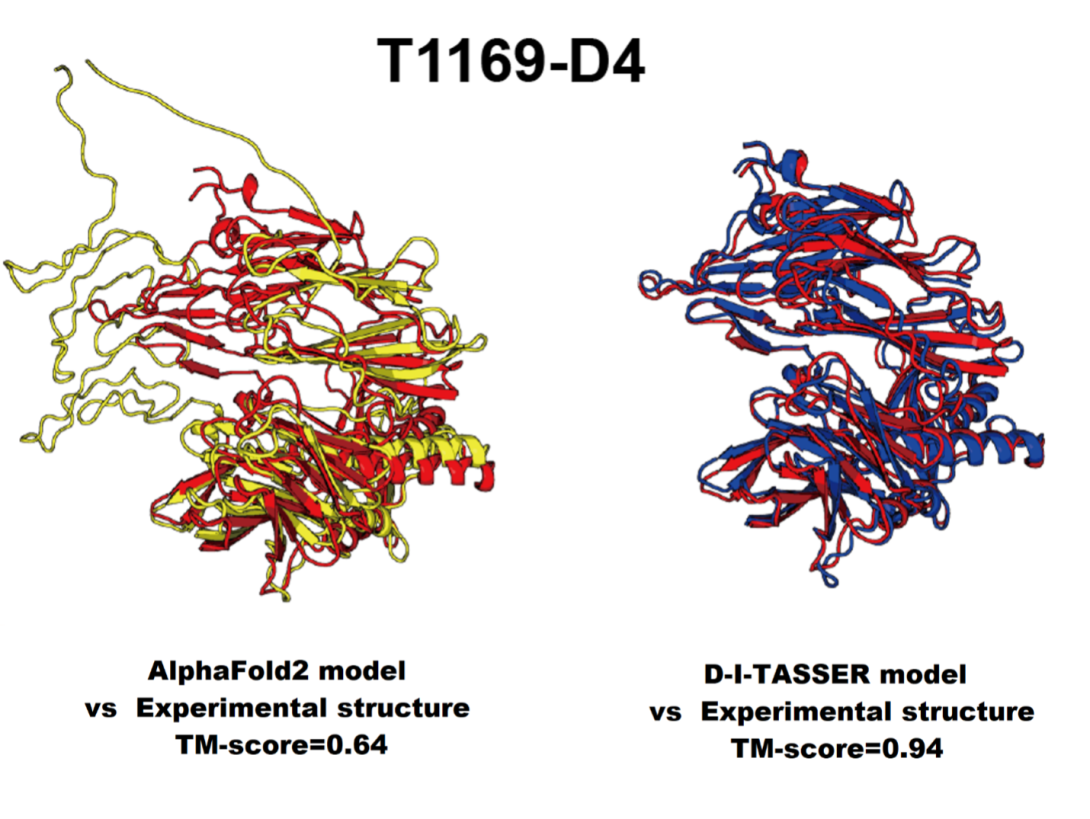
The Freddolino/Zhang/Zheng team competed against nearly 100 other groups which include other academic institutions, as well as major cloud and commercial companies, using their newly developed prediction programs D-I-TASSER and DMFold-Multimer. Groups from around the world submitted more than 53,000 models on 127 modeling targets in 5 prediction categories. Independent assessors then compared the models with experiments, and the results and their implications were discussed at the CASP15 Conference, held December 2022, in Antalya, Turkey.
“This is a highly competitive event, against some of the very best minds in the world,” said Freddolino. “We are very pleased with these results that place the University of Michigan at the leading-edge of computational and bioinformatics research again. In our CASP15 entries, we were able to take previous work from Dr. Zhang further, largely spearheaded by the work of Dr. Wei Zheng.”
These results were made possible in part thanks to the highly collaborative environment of the U-M, and the dedicated support from DCMB and several U-M Information and Technology Services (ITS) units and technical staff who gave priority to the project.
Brock Palen, Director of Advanced Research Computing (ARC), a division of ITS, provided monitoring and guidance on real-time impact and utilization of resources. “It was an honor to support this effort. It has always been ARC’s goal to take care of the technology so researchers can do what they do best. In this case, Freddolino and Zheng knocked it out of the park.”
“Having the flexibility and capacity provided by the Great Lakes High-Performance Computing Cluster was instrumental in meeting competition deadlines,” said Kenneth Weiss, IT project manager senior with DCMB and HITS. Jonathan Poisson, technical support manager with DCMB, also contributed significantly to this project by selecting and configuring the equipment purchased by the grant. “It was quite challenging to get these hardware components during the pandemic but these additional resources, added to the existing Great Lakes computational capacity, made this success possible,” said Poisson. “This assistance was crucial in meeting the tight CASP15 timelines, as each target is accompanied by a deadline for results,” said Weiss.
This research was supported by an NIH-funded grant (“High-Performance Computing Cluster for Biomedical Research,” SIG: S10OD026825).
The U-M team is now looking forward to the next CASP, in 2024!

Associate Professor