Dr. O'Meara leads the Maom lab, a computational pharmacology group that uses computational and statistical modeling to develop chemical probes and drugs. Congratulations Matthew!
Matthew O’Meara was promoted to Assistant Professor in the Department of Computational Medicine and Bioinformatics (DCMB), starting May 1, 2023. He joined the department as a Research Assistant Professor on September 1st, 2019.
Dr. O'Meara leads the Maom lab, a computational pharmacology group that uses computational and statistical modeling to develop chemical probes and drugs. They are interested in a wide range of biomedical and pharmaceutical projects, including drug repurposing to treat viral and microbial pathogens, and neuropharmacology.
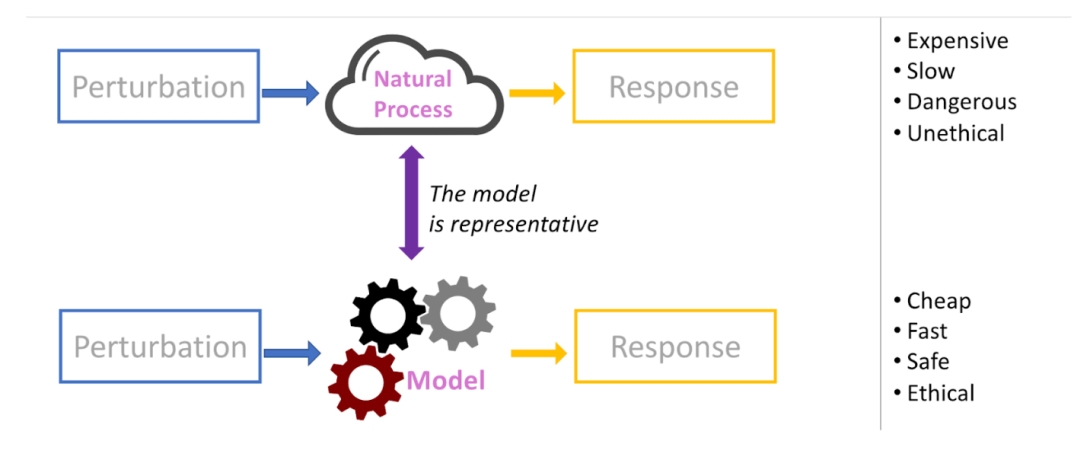
To better understand molecular interactions, the lab develops and applies two types of modeling. First, they use molecular simulations and deep neural networks to represent how molecules and chemicals interact. While computational models can be quite realistic, they are often intractable and difficult to compare with biochemical and phenotypic measurements. To address this issue, they then use statistical models to facilitate summarizing experimental data and making inferences. O’Meara’s research has been published in highly-recognized journals, including Science, Nature Communications and PNAS.
O’Meara’s modeling studies are very collaborative and his lab is contributing to many projects, analyzing the data and planning new experiments. DCMB brings together computational researchers from across the campus to understand how to apply computational approaches to biological problems. So, DCMB is the perfect fit to launch O’Meara’s computational pharmacology lab.
O’Meara is an active mentor and offers training to the modeling community, such as a Bayesian Statistical Modeling Workshop for Rosetta-based Modeling. He also participates in the Michigan Molecular Modeling group, and is the Co-director of an NSF funded Postbac program focused on training the next generation of researchers in biomolecular simulations.
Maom Lab website
Publication highlights include:
Gordon DE*, Jang GM*, Bouhaddou M*, Xu J, Obernier K*, White K*, O’Meara MJ*, et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature (2020)
Mirabelli C, Wotring JW, Zhang CJ, McCarty SM, Fursmidt R, Pretto CD, Qiao Y, Zhang Y, Frum T, Kadambi NS, Amin AT, O’Meara TR, Spence JR, Huang J, Alysandratos KD, Kotton DN, Handelman SK, Wobus CE, Weatherwax KJ, Mashour GA, O’Meara MJ*, Chinnaiyan AM*, Sexton JZ*, Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19, PNAS (2021)
Tummino TA, Rezelj VV, Fischer B, Fischer A, O’Meara MJ, Monel B, Vallet T, Zhang Z, Alon A, O’Donnell HR, Lyu J, Schadt H, White KM, Krogan NJ, Urban L, Shokat KM, Kruse AC, García-Sastre A, Schwartz O, Moretti F, Vignuzzi M, Pognan F, Shoichet BK, Phospholipidosis is a shared mechanism underlying the in vitro antiviral activity of many repurposed drugs against SARS-CoV-2, Science (2021)

Assistant Professor